Inverse designs#
In this notebook we will investigate a complex design created by topology optimization. The specific design is the Rasmus70nm.csv file from the RGB metalens problem of the photonics-opt-testbed repo.
import numpy as onp
import matplotlib.pyplot as plt
from skimage import measure
design = onp.genfromtxt("../../reference_designs/rgb_metalens/ex/Rasmus70nm.csv", delimiter=",")
design = design > 0.5 # Binarize
design = onp.rot90(design)
plt.figure(figsize=(7, 1))
ax = plt.subplot(111)
im = ax.imshow(1 - design, cmap="gray")
im.set_clim([-2, 1])
for c in measure.find_contours(design):
ax.plot(c[:, 1], c[:, 0], "k")
_ = ax.axis(False)
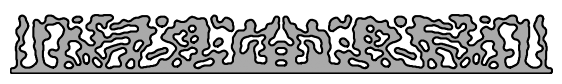
Now we’ll measure the length scale of solid and void features (i.e. the minimum width and spacing of features) using the imageruler.
import imageruler
min_width, min_spacing = imageruler.minimum_length_scale(design)
print(f"Minimum width is {min_width} pixels, minimum spacing is {min_spacing} pixels.")
Minimum width is 7 pixels, minimum spacing is 8 pixels.
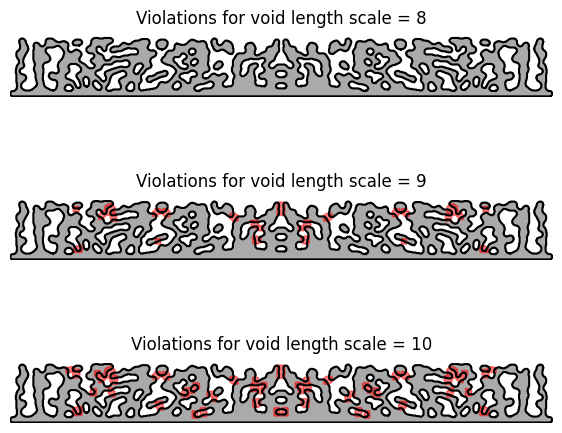
We find that the minimum width is 7 pixels, which means that we should find violations when checking for length scales greater than 7 pixels. We can identify these violations using the length_scale_violations_solid function:
from scipy import ndimage
plt.figure(figsize=(7, 6))
for i, dim in enumerate(range(min_width, min_width + 3)):
width_violations = imageruler.length_scale_violations_solid(
design, length_scale=dim
)
width_violations = ndimage.binary_dilation(width_violations, onp.ones((10, 10)))
ax = plt.subplot(3, 1, i + 1)
im = ax.imshow(1 - design, cmap="gray")
im.set_clim([-2, 1])
for c in measure.find_contours(design):
ax.plot(c[:, 1], c[:, 0], "k")
for c in measure.find_contours(width_violations):
ax.fill(c[:, 1], c[:, 0], "r", alpha=0.5, ec="r")
ax.set_title(f"Violations for solid length scale = {dim}")
ax.axis(False)

And similarly, we can find void length scale violations:
plt.figure(figsize=(7, 6))
for i, dim in enumerate(range(min_spacing, min_spacing + 3)):
width_violations = imageruler.length_scale_violations_solid(
~design, length_scale=dim
)
width_violations = ndimage.binary_dilation(width_violations, onp.ones((10, 10)))
ax = plt.subplot(3, 1, i + 1)
im = ax.imshow(1 - design, cmap="gray")
im.set_clim([-2, 1])
for c in measure.find_contours(design):
ax.plot(c[:, 1], c[:, 0], "k")
for c in measure.find_contours(width_violations):
ax.fill(c[:, 1], c[:, 0], "r", alpha=0.5, ec="r")
ax.set_title(f"Violations for void length scale = {dim}")
ax.axis(False)